Biomarkers: Promising Tools for Detection of Early- Stage-Lung Cancer
Lucia Kiio* , Damaris Mbui and Peter Ndangili
Lucia Kiio*
University of Nairobi, Kenya
- *Corresponding Author:
- Lucia Kiio
University of Nairobi, Kenya
E-mail: lucy.k630@gmail.com;
lkiio@students.uonbi.ac.ke
Received Date: May 28, 2021; Accepted Date: June 14, 2021; Published Date: June 21, 2021
Citation: Kiio L (2021) Biomarkers: Promising Tools for Detection of Early- Stage-Lung Cancer. Med Clin Rev Vol.7 No.6:140.
Abstract
Lung cancer, a malignant lung tumor with several histological variants arising from various cell types for instant alveoli, bronchial mucous gland, bronchioles, and bronchial epithelium, continues to cause significant mortality worldwide. As advances in therapy treatment continue to be made, the overall prognosis for the patients diagnosed with the tumor remains poor. While the prognosis of lung cancer is generally bleak, with 5-year survival rates of only 15%, there is hope, and evidence, that early detection of lung cancer can reduce the mortality rate significantly. Traditionally, markers such as age, status, performance, and disease stage have been used to risk-stratify patients and also guide therapeutic decisions. Useful information is obtained from these markers, but more sensitive markers are urgently needed. For several years, researchers throughout the world have extensively investigated several screening modalities for lung cancer detection, such as computerized tomography, chest X-ray, positron emission tomography, sputum cytology, magnetic resonance imaging, and biopsy. However, these techniques are not convenient for patients with other pathologies, they have also been found to produce high cost and potentially false- positive results. Developing a rapid and sensitive technique for the early diagnosis of lung cancer is needed. Several markers have been identified by molecular and genetic studies, which seem to play critical roles in carcinogenesis and influence the outcome of the patient. This article reviews statistics of lung cancer in general, types of lung cancer, several biomarkers that have been identified in lung cancer, their predictive and prognostic roles. This review also summarizes the most recent potential biomarkers that have been discovered, some biomarkers and biosensors used in the detection of cancer, their detection limits, and challenges and future prospective.
Keywords
Biomarkers; Biosensors; Early diagnosis; Lung cancer
Introduction
Cancer can be defined as abnormal and uncontrolled cell growth in the biological system. Due to this uncontrolled growth, an unregulated tumor mass forms. The cells become resistant to apoptosis and spread and metastasize to other parts of the body, beyond the place of origin, rendering the cancer incurable. It may be caused by accumulated specific genetic and epigenetic effects, which could have their origin as environmental sources, hereditary sources, or both [1].
According to global statistics by Bray et al., there was an estimated 18.1 million new cancer cases (17.0 million excluding nonmelanoma skin cancer) and 9.6 million cancer deaths (9.5 million excluding nonmelanoma skin cancer) in 2018 [2]. In both males and females combined, lung cancer is the most commonly diagnosed cancer with 11.6% of the total cases and has the highest death rate of 18.4% of the total cancer deaths, closely followed by female breast cancer (11.6%), prostate cancer (7.1%), and colorectal cancer (6.1%) for incidence and colorectal cancer (9.2%), stomach cancer (8.2%), and liver cancer (8.2%) for mortality [2]. Among males, lung cancer is the most frequent cancer and the leading cause of cancer death, followed by prostate and then colorectal cancer (for incidence) and liver and stomach cancer (for mortality) [3]. Among females, breast cancer is the most frequently diagnosed cancer and the leading cause of cancer death, followed by colorectal and lung cancer (for incidence), and vice versa (for mortality); cervical cancer ranks fourth for both incidence and mortality [3]. The most commonly diagnosed cancer and the leading cause of cancer death, however, extensively differ across countries and within each country depending on the level of economic development and associated social and lifestyle factors [2].
Lung Cancer
Lung cancer has currently become a global predicament, from a subtle disease to an emerging public health issue. Current studies show that this disease has recorded the highest mortality among all types of cancer [4]. The etiological factors of lung cancer have become more diverse because of increasing industrialization and environmental pollution around the world [4].
The global cancer statistics 2018 showed that lung cancer had the highest morbidity (11.6 percent) of total cases and mortality (18.4 percent of total cancer deaths) in the world [2]. Lung cancer has a 5- year survival rate as low as 15% [5]. When the cancer is discovered early, the survival rate can be up to 70% for small, localized tumors (stage I). However, many patients (approx. 75%) usually have advanced disease at the time of diagnosis (stage III/IV) and despite significant developments in the oncological management of late-stage lung cancer over recent years, survival remains poor [6]. Despite the low lung cancer mortality rate, the majority of patients are still diagnosed with advanced or metastatic lung cancer, leading to poor outcomes [6]. Additionally, even though there are advances in the treatment of lung cancer, the prognosis of patients is not gratifying [6]. Detection of lung cancer in the early stages before clinical symptoms can be an effective means to reduce the lung cancer mortality rate. Therefore, it would be important to find effective and reliable biomarkers to diagnose lung cancer early, before the tumor cells metastasize and the symptoms of cancer become evident. An efficient and effective means of early lung cancer prognosis can be by use of biomarkers, which may curtail the recurrence of lung cancer after surgery and improve the life of patients.
Types of Lung Cancer
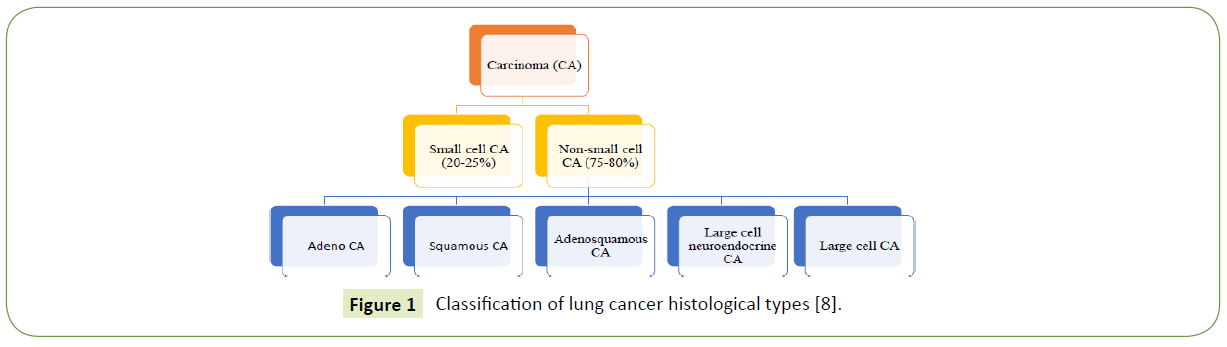
Lung cancer is a malignant lung tumor that may arise from the bronchial epithelium, bronchioles, alveoli, or bronchial mucous glands [7]. It is characterized by post-treatment relapses, metastasis, and a range of histological types [7]. In 2015, the World Health Organization (WHO) classified lung tumors into two [8], namely the small-cell lung carcinoma (SCLC) and non-smallcell lung carcinoma (NSCLC) (Figure 1).
Figure 1 Classification of lung cancer histological types [8].
Small-cell lung cancer (SCLC), is a high-grade tumor with specific histological and genetic characteristics [9]. It is characterized by small size cells, absence of differentiation, fast tumor growth, metastasis at early stages, and release of specific biomarkers and hormones [9]. On the other hand, Non-small cell lung carcinoma (NSCLC) is any type of epithelial lung cancer other than small cell lung carcinoma and accounts for approximately 85% of all lung cancers. NSCLCs are mainly treated by surgical resection, although chemotherapy (i.e., adjuvant chemotherapy) is increasingly being used [10]. NSCLC is classified in to adenocarcinoma, adenosquamous carcinoma, squamous cell carcinoma, large cell carcinoma, and large cell neuroendocrine carcinoma [8]
Origin and Historical Features of Lung Cancer Types
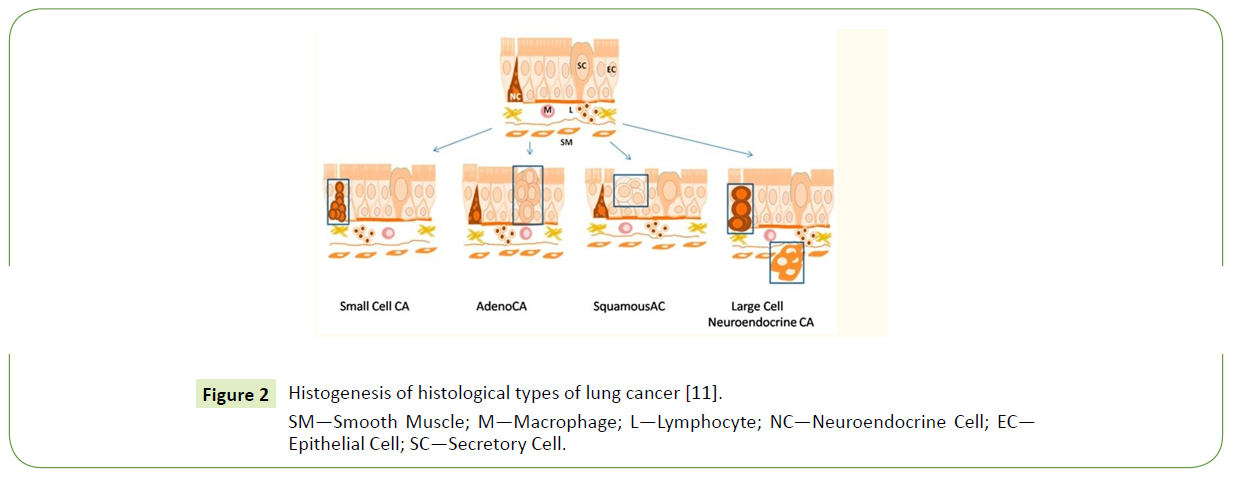
There are two viewpoints on the origin of Small-cell lung cancer (SCLC). According to the first hypothesis, SCLC arises from cells of the diffuse endocrine system, i.e., the Amine precursor uptake decarboxylation (APUD)-system (Figure 2), the second suggests this type of lung cancer originates from the endoderm bronchial lining layer [7].
SM—Smooth Muscle; M—Macrophage; L—Lymphocyte; NC—Neuroendocrine Cell; EC— Epithelial Cell; SC—Secretory Cell.
Figure 2 Histogenesis of histological types of lung cancer [11].
As can be seen in Figure 2, among the other subtypes of non-small- cell lung carcinoma (NSCLC), adenocarcinoma emanates from glandular cells of the bronchial mucosa and now serves as the dominant histological subtype among the other lung cancer types. Squamous lung cancer originates from the modified bronchial epithelial cells and is characterized by keratinization, keratin pearl formation, or the presence of intercellular bridges.
Аdenosquamous carcinoma is a type of cancer that contains two types of cells: squamous cells (thin, flat cells that line certain organs) and gland-like cells [12]. Large cell neuroendocrine carcinoma is a malignant epithelial tumor, which consists of large polygonal cells that do not show any obvious evidence of histological differentiation. The cases include large cell neuroendocrine carcinoma, basaloid carcinoma, lymphoepithelioma-like carcinoma, and clear cell carcinoma. The tumor arises from neuroendocrine cells of the respiratory tract lining layer or smooth muscle cells of its wall, Figure 2. A Large-cell carcinoma is a heterogeneous group of undifferentiated malignant neoplasms that lack the cytologic and architectural features of small cell carcinoma and glandular or squamous differentiation. Large-cell carcinoma is categorized as a subtype of NSCLC that arises from epithelial cells of the lung [11].
It is well known that a unique combination of exogenous and endogenous factors influences the occurrence and development of lung cancer in each individual. Lung cancer, like any other oncological disease, is heterogeneous. Therefore, in addition to various histological types, this disease has many molecular and pathological subtypes characterized by heterogeneous cellular genetic and epigenetic changes and a different combination of protein biomarkers. At present, data on protein signatures of molecular subtypes of histological types of lung cancer is extremely limited, though a large number of genetic studies reflecting the probability of certain mutations in genes are presented. In particular, mutations of the epidermal growth factor receptor (EGFR) in lung adenocarcinoma have been well studied. It was found that in patients with lung adenocarcinoma, the probability of EGFR mutations increases linearly from 3.7% (18–30 years) to 18.5% (81–100 years), and in female nonsmokers, the probability of mutations is higher than in men [13]. In male non-smokers, the probability of EGFR mutation is much higher than in smokers [14].
Identification of the correct histological type of lung cancer and their molecular subtypes is necessary due to the different available treatment strategies. Tumor cells of each histological type release certain protein biomarkers into the bloodstream and therefore play a key role in cancer genesis. The use of blood plasma to determine the origin and nature of the malignant cells for diagnosis requires knowledge about the expression of protein biomarkers, their specificity, sensitivity, and their release by different types of lung cancer cells [15, 16].
Small Cell Lung Cancer
SCLC originates from neuroendocrine cells of the APUD-system (amine precursor uptake and decarboxylation system) [17]. SCLC has two of the main biological features of these cells. These features include the production of L-3,4-dihydroxyphenylalani nedecarboxylase (L-DOPA- decarboxylase) and the production of neuron-specific enolase (NSE). L-DOPA decarboxylase is the gene encoding for the enzyme that catalyzes the biosynthesis of dopamine in humans [18]. NSE is a glycolytic neuron-specific enzyme of enolase with two almost identical 39-kDa polypeptides produced in the central and peripheral neurons and malignant tumors of neuroectodermal origin.
NSE is localized to neurons and neuroendocrine cells of the amine precursor uptake and decarboxylation (APUD) series. It is found in various neuroendocrine origin or neuronal tumors such as SCLC and neuroblastoma [19]. Adrenocorticotropic hormone, serotonin, antidiuretic hormone, calcitonin, growth hormone, melanocyte-stimulating hormone, and estrogen are also produced in SCLC [20].
The other well-known biomarker of SCLC is a pro-gastrin-releasing peptide (ProGRP) [21]. High levels of ProGRP were found in the blood of patients with SCLC and medullary thyroid cancer (>200 pgmL−1). Blood plasma of healthy people and patients with benign diseases have ProGRP concentrations of 35 pgmL−1 and 4.5 x 103 pgmL−1 respectively. ProGRP has organ specificity and does not correlate with the stage of lung cancer. Although ProGRP is more specific than NSE, its use in biosensors is complicated due to its instability and difficulty of identification [21]. The sensitivity and specificity of ProGRP were 80% and 90%, respectively, while that for NSE was 64% and 43%. However, 27% of patients with SCLC had increased levels of NSE and normal levels of ProGRP.
Classification of non-small-cell lung carcinoma (NSCLC)
NSCLC is divided into five subtypes which include; Squamous Lung Cancer, Adenocarcinoma, Large Cell Carcinoma, Аdenosquamous Carcinoma and Large Cell Neuroendocrine Carcinoma.
Squamous Lung Cancer
Squamous lung cancer originates from modified bronchial epithelial cells. One of the most distinctive features of squamous lung cancer is high levels of fragmented cytokeratin CK-19 subunit, CYFRA 21-1. CK-19 is a protein component of intermediate fibers of epithelial cells [22]. The level of CYFRA 21-1 is increased during the malignization process of normal epithelial cells. CYFRA 21-1 is highly expressed in the serum of patients with a metastatic form of squamous lung cancer [23].
The other specific protein for squamous lung cancer is squamous cell carcinoma antigen (SCCA), a 48-kDa protein which is found in increased levels in squamous lung cancer [24]. SCCA is an inhibitor of serine proteases such as human chymotrypsin-like elastase (CELA), calpain 1 (CAPN1), and cathepsin L (CTSL) [25]. It also inhibits apoptosis of tumor cells and stimulates invasion and metastasis [25].
Adenocarcinoma
Adenocarcinoma is another type of lung cancer that arises from glandular cells of the bronchial mucosa and expresses several protein markers.Diagnosis of adenocarcinoma is often based on the identification of molecular markers of mutations, in particular EGFR, Excision repair cross-complement (ERCC), ribonucleoside-diphosphate reductase (RRM 1), KRAS proto-oncogene (KRAS), thymidylate synthetase (TS), and echinoderm microtubule-associated protein-like 4 gene fused to anaplastic lymphoma kinase receptor tyrosine kinase (EML4-Alk) (Sholl 2015). Recently, protein PSF3 (DNA replication complex GINS) has become popular as a biomarker of adenocarcinoma. PSF3 is a member of the heterotetrameric complex GINS (“go-ichi-ni-san” complex, from the first letters of the Japanese numbers 5-1-2-3) comprising Systemic RNA interference defective protein 5 (SLD5), GINS complex subunit 1 (PSF1), GINS complex subunit 2 (PSF2), and GINS complex subunit 3 (PSF3). This complex associates with proteins, which in turn regulate both the initiation and the progression of DNA replication [26]. To date, an overexpression of PSF3 in adenocarcinoma has been established, which leads us to conclude that its level should be higher in blood plasma. However, data on the level of PSF3 in the blood is yet to be reported. In addition to these biomarkers, several novel lung adenocarcinomas associated proteins have been found using aptamers, such as lamin (LMN) and vimentin (VIM)[27], neutrophil defensin (DEF), and tubulin (TUB), cytoplasmic actin (ACT), cathepsin D (CTSD), clusterin (CLU), nucleolin (NCL), and mucin-1 (MUC1). According to recent studies, the identification of such proteins would improve the diagnosis of adenocarcinoma.
Large Cell Carcinoma
Large cell carcinoma is a malignant epithelial tumor that comprises large polygonal cells showing no obvious evidence of histological differentiation. Large cell carcinoma is characterized by small, scattered groups of large non-differentiated, polymorphic, and often dual- or multi-core cells [28].
Аdenosquamous Carcinoma
Аdenosquamous carcinoma is characterized by the features of squamous cell carcinoma and adenocarcinoma simultaneously. Therefore, it has a protein biomarker of both histotypes.
Large Cell Neuroendocrine Carcinoma
Large cell neuroendocrine carcinoma (LCNEC) is extremely rare. There are difficulties related to its diagnosis and treatment.
LCNEC shows over expression of topoisomerases somatostatin precursor (TOP SST), and excision repair 1, endonuclease non-catalytic subunit (ERCC1).
Biomarkers
Biomarkers can be defined as simply structural molecules that represent the biological homeostasis and thus make it possible to detect changes in physiology. More specifically in the case of cancer, tumor markers indicate the presence of malignant cell growth when they are measured in higher concentrations than usual. An ideal biomarker should have the following characteristics: a) It should be highly sensitive: its concentration should increase in the presence of the disease; b) It should be highly specific: it should not express in the absence of the disease; c)It should change in accordance with clinical evolution, reflecting the current status of the disease; d) It should show the presence of relapse before symptoms reappear at the clinical level; e) It should be reproducible; and f) It should be relatively easy and have a low cost to measure in the sample [29,30].
Biomarkers for lung cancer detection
Lung cancer biomarkers would help to differentiate between cancerous and normal cells. These biomarkers can be used potentially to develop a more effective diagnostic tool for lung cancer. Body fluids such as pleural effusions, blood, urine, etc. that are in contact with the cancerous cells are enriched with proteins shed from the tumors. These proteins shed from the cancer cells can enter the blood circulation and can be monitored in body fluids.
Classification of Cancer Biomarkers
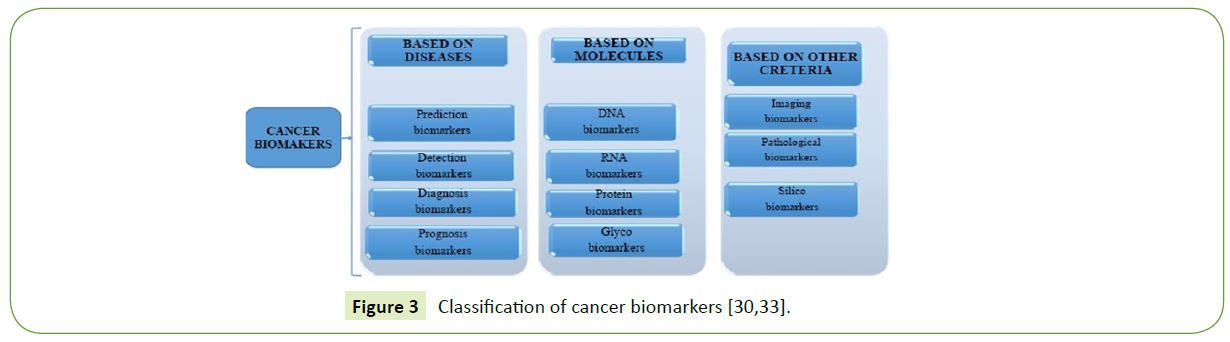
Cancer biomarkers can play a significant role in early diagnosis and prognosis. According to the past several decades’ research, cancer biomarkers can be classified into four types based on the disease state, as represented in Figure 3.
The first type is the diagnostic or screening biomarkers. These are used to detect and identify an explicit type of cancer. To be effective, this sort of biomarker should be very specific and sensitive [31]. Secondly, there are prognostic biomarkers, which are utilized once the disease status has been determined, to foresee the expected evolution of cancer. The third type is the predictive or stratification biomarkers. These are applied to predict the response to treatment and finally, there are the detection biomarkers [32].
Biomarkers are also classified based on biomolecules and other criteria such as imaging, pathological and silico biomarkers as shown in Figure 3. Biomarkers can also be classified based on the different areas they work in. For instance, a distinction can be made between genetic, epigenetic, proteomic, metabolic, and micro RNA-related biomarkers. The growth of a tumor is associated with both genetic alterations, and the release of proteins, metabolites, nucleic acids and other biomolecules.
Genetic-based biomarkers are detected using DNA arrays, polymerase chain reaction (PCR), reverse transcriptionpolymerase chain reaction (RT-PCR), DNA sequencing, or fluorescent in situ hybridization (FISH). For the determination of the levels of certain protein biomarkers, proteomic techniques are applied. These include mass spectrometry, enzyme-linked immunosorbent assay (ELISA), and immunohistochemistry. Metabolites are regularly detected using liquid chromatography [34,35].
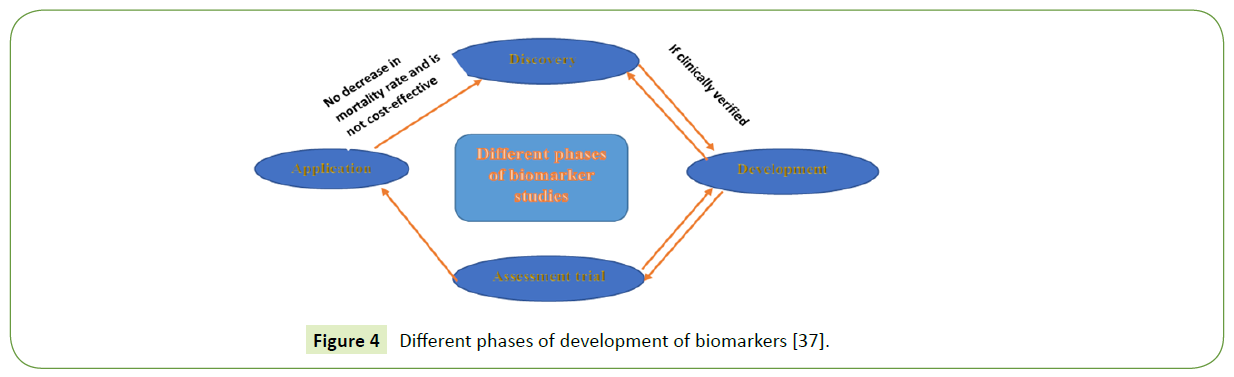
Proteomics has assisted in the diagnosis of protein-based biomarkers; nonetheless, the development of such biomarkers requires a systematic approach and critical evaluation before its progression from laboratory to clinic [36] Figure 4.
Figure 4 Different phases of development of biomarkers [37].
A specificity of 100% is required in clinical terms for biomarkers [38], but may not be always practical as it may become less sensitive. Several investigators thus use a specificity of > 85% as the cut-off point [39]. The link between sensitivity and specificity for a certain disease under prediction assigns suitable cut-off points, which give rise to a receiver operating characteristic (ROC) curve. A clinically important biomarker would be the one with the largest area under the ROC curve. Taking 100% specificity as a reference point, such ROC curves aid in calculating higher sensitivity for a biomarker, hence reducing the burden of false-positive cases by 5% [40].
Several important biomarkers investigated for lung cancer diagnosis are listed in Table 1. Some of them are already in use, while others have shown considerable potential and are still being studied. The most broadly applied division is the one between DNA- or genetic-based biomarkers and protein or proteomics-based biomarkers. Examples of DNA and proteinbased biomarkers are as shown in Table 1.
| Genetic biomarkers | RAR-βmRNA,COX2,DAPK,RASSF1A,IL-8mRNA,FHIT, K-rasmutant,p53 mutant,EGFR |
| Protein biomarkers | CEA, CYFRA21-1, TPA, tumor M2-pyruvatekinase,haptoglobin-α2,APOA1,KLKB1,ProGRP,α-enolase(NSE),α-1-acidglycoprotein,chromograninA,bombesin-likegastrin-releasingpeptide, CK-BB, cytokeratin-7, CA 19-9,CA125,plasmakallikreinB1,VEGF,nitratedceruloplasmin, annexinII,CD59glycoprotein,transthyretin(TTR),GM2AP |
Table 1 DNA- and protein-based biomarkers in lung cancer detection [24].
For lung cancer diagnosis, both genetic and protein markers are examined. However, none of the DNA tumor markers investigated so far seem to be sensitive and selective enough to be broadly used as a diagnostic tool. One of the reasons for this is that the mRNA levels are not linked directly to the protein levels. The advantage of protein tumor markers is that they can be more specific to the cancer type and status because they show more variety. Diversity is created for example by post-translational modifications (such as acetylations, glycosylations, phosphorylations, and methylations), protease cleavages, or alternative splicing [41,42].
The level of protein biomarkers can be determined in several body fluids, such as blood, serum, sputum, cerebral spinal fluid, and urine. The fact that much information can be obtained non-invasively is a big asset of proteomics. In genetics, on the other hand, the DNA usually has to be extracted from the cells before analysis is possible. Often, an extra enrichment is necessary because the concentration of the markers generally is very low. Tests have also been performed on the determination of certain oncomarkers in exhaled breath [43]. In this review, protein biomarkers such as Carcinoembryonic Antigen (CEA), Cytokeratin fragment 21-1(CYFRA21-1), Neuron-specific enolase (NSE), Tumor M2- pyruvate, C- reactive protein (CRP), and the currently used biomarkers such as Exosomal biomarkers will be reviewed as promising tools for early detection of lung cancer.
Carcinoembryonic Antigen (CEA)
Carcinoembryonic antigen is a serum protein that can be used as a diagnostic or therapeutic tumor marker in different types of cancer. It is a glycoprotein, belonging to the immunoglobulin superfamily. The human CEA is encoded by a gene located on chromosome 19 and has a molecular weight of about 70 kD, which can run up to 200 kD through glycosylations. It is an oncofetal protein that is not usually expressed in adult tissues. The CEA levels in the blood of lung cancer patients are usually elevated and they are inversely correlated with the response to cancer therapy. Thus CEA biomarker is used to detect cancer reoccurrence and also a prediction of poor survival rate [44].
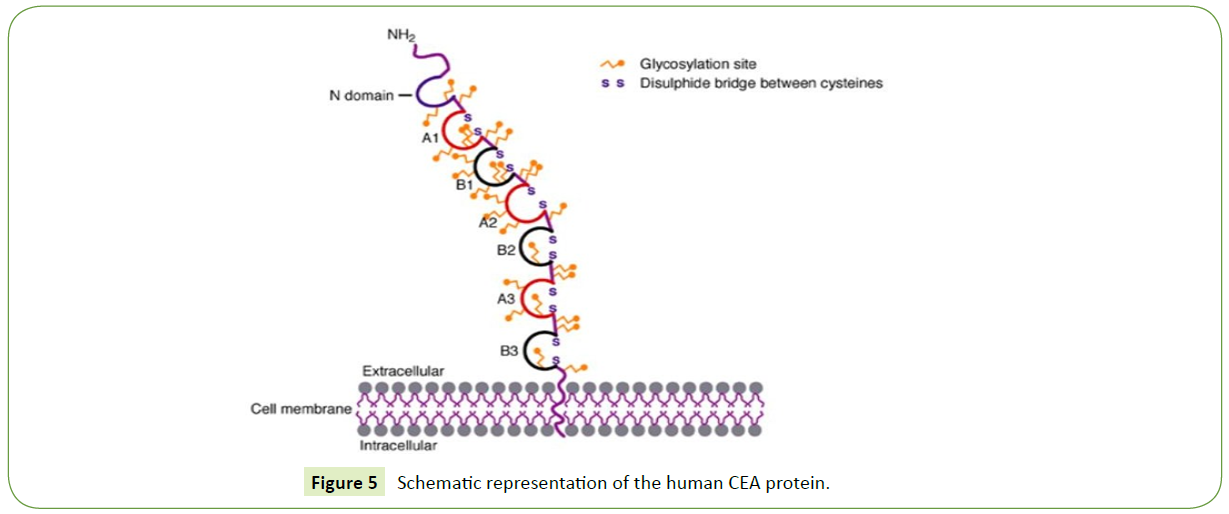
The CEA protein consists of an N-terminal sequence, three repeated sulphide-linked domains comprising 178 amino acids each, and a C-terminal region. Every repeated domain can be split up into two subdomains, which show sequence similarities. The C-terminus is hydrophobic and is associated with the cell membrane, differentiating CEA from other members of the CEA family, such as the secreted PSGs (pregnancy-specific glycoproteins). The structure of the CEA protein is illustrated in Figure 5.
CEA was first mentioned as a tumor-related antigen by Gold and Freedman in 1965. In the past decades, many clinical assays have been performed on this potential tumor marker. In contrast to what was believed earlier, it is now clear that CEA is not only expressed in fetuses and cancer patients but also healthy adults. A normal adult who does not smoke generally has levels of CEA that are below 2.5 ng/mL. The concentration is higher in smokers, but is commonly less than 5.0 ng/mL, while in cancer patients, it can rise to above 100 ng/mL. Elevated levels of the oncofetal glycoprotein are mainly seen in the serum of people with colorectal, breast, and lung carcinomas [45,46].
Since an increased level of CEA can be an indication of the growth or recurrence of a tumor, the detection of the elevated amount of CEA in the serum could be of use in the diagnosis of cancer. However, one of the main restrictions for the application of CEA as a diagnostic screening tool for lung cancer is that the expression of this glycoprotein is also increased in other, non-malignant diseases, such as bronchitis, cirrhosis, hepatitis, inflammatory bowel disease, pancreatitis, etc. The levels of CEA in benign conditions normally do not exceed 10 ng/mL [47]. Research has also been done on the determination of tumor markers in pleural effusions [48].
Zhang et al., noted that the levels of CEA are significantly higher in patients with lung cancer than in people with benign pleural effusions [49].
Cytokeratin fragment 21-1 (CYFRA21-1)
Cytokeratins are structural proteins forming the subunits of epithelial intermediary filaments. Fragments of cytokeratin-19 (CYFRA 21-1) are frequently released into the blood of cancer patients. Due to the high specificity relative to benign lung diseases, it is suitable for use as a differentiation marker in tumor pathology especially in lung cancer [50]. Cytokeratins are intermediate filaments of the cytoskeleton that are specifically expressed in epithelial cells, considered to be protein markers for the detection of epithelial tumor cells [51]. CYFRA21-1 protein has been recognized as a very sensitive biomarker to identify NSCLC, especially squamous cell carcinoma [52]. Previous research findings have confirmed the sensitivity and specificity of CYFRA21-1 as a tumor marker [53]. CYFRA21-1 gene has been reported as the most robust DNA-based biomarker for NSCLC [54]. Subjects with advanced NSCLC observed high-level serum CYFRA21-1 [55], and therefore high serum CYFRA21-1 levels may be useful non- invasive marker to identify NSCLC risk.
Neuron-specific enolase (NSE)
Neuron-specific enolase (NSE) is a highly acidic protein that was first isolated from nervous tissue in 1965 by scholars who were investigating nervous system-specific proteins using DEAE-cellulose column chromatography and starch gel electrophoresis. NSE is a key enzyme of the glycolytic pathway and is generally located in the cytoplasm of neurons and, characteristically, neuroendocrine cells. NSE leaks out of neurons during necrosis, so it is also an important marker of neuronal damage [56].
Studies show that serum NSE levels are significantly higher in advanced lung cancer patients than in benign lung disease patients [17]. Serum NSE levels also are significantly higher in SCLC cases than other tumor pathologies, including adenocarcinoma and squamous cell carcinoma. Studies also show that serum NSE levels significantly correlate with the prognosis of advanced lung cancer patients and suggest that NSE can be used as a secondary indicator in prognosis prediction, especially for SCLC [17].
Tumor M2- pyruvate
Tumor M2- pyruvate kinase (PKM2) is a dimeric isoenzyme of pyruvate kinase type M2 that is present as an active dimer and less active tetramer.
PKM2 is increased in various cancers, for instance, the dimeric isoform is typical of tumor cells and its levels can be measured in body fluids like blood. According to Rzechonek et al., Bandyopadhayaya & Mandal, PKM2 has a specificity of 50% and sensitivity of 79% in patients with SCLC while patients with NSCLC have specificity and sensitivity of 81% [57,58]. These levels correlate well with tumor progression and remission making it a very useful tool for disease monitoring.
C- reactive protein (CRP)
C- reactive protein (CRP), a well-known member of the pentraxin family, has a high affinity for diverse types of extrinsic and autologous ligands [59]. Autologous ligands comprise Fc-γ receptors, phospholipids, damaged cell membranes, apoptotic cells, plasma lipoproteins, ribonucleoprotein particles, and extracellular matrix proteins [59]. On the other hand, extrinsic ligands consist of various constituents of several microorganisms. Once aggregated or constrained to macromolecular ligands, CRP stimulates the complement system to form a membrane attack complex, which thereafter attacks target molecules or cells. Thus, CRP plays an important role in the clearance of abnormal or apoptotic cells [60,61].
CRP is secreted by hepatocytes in response to the inflammatory cytokines produced by the tumor microenvironment [62]. It can enter the tumor microenvironment through the circulation, where it binds to a variety of autologous and extrinsic ligands and plays a key role in the clearance of tumor cells [60].
As CRP can bind to various ligands, a subset of the circulating CRP pool may exist in the form of CRP complexes. Because elevated circulating levels of CRP have been frequently found in lung cancer patients, CRP may bind to ligands expressed by lung cancer cells [63] or tumor-associated cells, and that CRP-bound components in the serum of lung cancer patients may be different from those of healthy subjects. Therefore, CRP-bound complexes in the serum may be potential prognostic biomarkers of lung cancer.
D- reactive protein (CRP), a routinely measured marker of chronic inflammation, is a reliable biomarker for perioperative management because it can aid in the early detection of surgical site infection. Furthermore, CRP is an acute reactant protein that is increasingly expressed in the presence of infection, trauma, tissue necrosis, tumor, and several types of inflammatory diseases. CRP is an important prognostic indicator in patients with various malignancies, such as pancreatic [64,65], urological [66] hepatocellular [67], and colorectal cancers [68] and [69]. Furthermore, some studies [70,71] showed that elevated serum CRP levels may predict poor survival in patients with NSCLC. Nonetheless, other studies demonstrated that elevated CRP levels are unrelated to low survival [72,73].
Exosomal biomarkers
According to Wu & Shen, 2020, Exosomes are vesicles with a diameter of 40‐100 nm [74]. They emerge to form early multivesicular bodies (MVB). When joined with the plasma membrane, they create an intracellular vesicle (ILV), which is released into the extracellular environment [75]. It is evident that majority of cells can secrete exosomes. Therefore, exosomes are extensively present in several biological fluids, including urine [76], plasma [77], saliva [78], bile [79], semen, breast milk [80], amniotic fluid [81], serum [82] and cerebrospinal fluid [83]. Separation of Exosomes can be done using; ultracentrifugation [84], precipitation [85] and microfluidic chip [86]. Exosomes can be extracted from human body fluids using various separation techniques, and the number of exosomes in plasma and serum samples is consistent, [87]. The number of bronchoalveolar lavage (BAL) exosomes is less than the number of plasma exosomes, [88]. In comparison with BAL samples and serum samples, plasma exosomes obtained by ultracentrifugation contain more biomarkers [88].
In addition, more and more evidence indicates that an increase in unregulated secretion of exosomes in cancer cells are associated with tumorigenesis [89]. Exosomal miRNAs biomarkers play a significant role in several cancers, including; lung cancer, nasopharyngeal carcinoma [90], and colorectal cancer [90]. Exosomes can also be used as a liquid biomarker for the diagnosis, prognosis, and treatment of head and neck squamous cell carcinoma [91], however, there is no evidence that it can be used to relate Barrett’s esophagus and esophageal adenocarcinoma patients with other subjects differentiating circulating exosomal miRNAs [92]. The current studies indicate that exosomes are closely associated with the occurrence of lung cancer. Furthermore, they show that tumor‐derived exosomes can be involved in the occurrence and development of lung cancer by regulating multiple pathways, for instance; engaging in epithelial‐mesenchymal transformation (EMT) [93] reinforcing tumor angiogenesis and vascular permeability [94] and boosting chemotherapy resistance [95].
Lately, progressive studies have presented that exosomal miRNAs are current biomarkers of lung cancer. Exosomal miRNAs have been spotted in several human body fluids and are used as noninvasive biomarkers for carcinoma detection [96]. Exosomes act as a span for information exchange amid cells, which can transport miRNAs and hinder RNA enzyme degeneration. Exosomes can as well influence cell‐cell communication by ferrying their contents to target cells in the lung cancer microenvironment [97], thus exosomes can be used as quintessential liquid biopsy specimens.
According to Dejima et al., 2017, 12 specific miRNAs were proved to be inflated in NSCLC and were reflected in circulating exosomes when circulating tumor exosome levels in plasma samples from 28 patients with lung adenocarcinoma were compared with 9 healthy controls [98]. The level of exosomal miRNAs in body fluids of lung cancer patients was up‐regulated, which showed that exosomal miRNAs played an important role in the development and progression of lung cancer.
Wu et al., found that the detection ability of serum exosomal miR‐216b was better than CEA, CYFRA21‐1, and SCCA, and the combination of serum exosomal miR‐216b and CEA, CYFRA21‐1 and, SCCA produced an Area Under Curve (AUC) value from 0.84 to 0.925 [74]. Table 2 show some of the biomarkers and biosensors used in the detection of cancer and their detection limits.
| Biomarker | Biosensor | Surface Ligand | Detection Limit | Line arrange | Reference |
|---|---|---|---|---|---|
| AFP | Electrochemical* | Microfluidic immunosensor | 3.03 pgmL−1 | 0.01 ng mL−1 -50ngmL−1. | [99] |
| Graphene and thioninenano composite film |
0.050 μg/mL | 0.1–100.0 μg/mL | [82] | ||
| CEA | Electrochemical* | Antibody | 9.4ng/mL. | - | [100] |
| SPR* | Antibody | 3.05fgmL−1 | 1.0×10−14gmL−1- 1.0×10−10gmL−1 |
[101] | |
| SPR** | 17.8pg/ml | - | [102] | ||
| CRP | QCM* | Antibody | 0.1μg/mL | 0.1μg/mLto1 mg/mL |
[103] |
| DNA mutations |
SPR* | ssDNA | 0.0028nM | - | [104] |
| CAI5-3 | Electrochemical* | Antibody | 15 × 10−6U/mL | 50–15 × 10−6U/mL | [105] |
| AFP | SPR** | Antibody | 0.1 ng/mL | - | [106] |
| CRP | Electrochemical* | Antibody | 0.025ngmL−1 | 0.01–10,000 ngmL−1 | [107] |
| TNF | Electrochemical* | Antibody | 97.2pM | 0- 22.3nM | [108] |
| EGFR | Optical* | Aptamer | 20 nM | - | [109] |
| Lab-on-a-chip* | Antibody | 0.01 pg mL−1 | 0.05–50000pg mL−1 | [110] | |
| K-ras pointmutation | SPR** | PNA | 16fM | 50fM-1nM | [77] |
| PSA | Electrochemical* | Nanoparticle- antibody |
0.2and 0.07ng/mL |
- | [111] |
| SPR* | Antibody | 18×10−14 M | 0.1–50ngmL−1 | [112] | |
| p53gene | SPR** | Antibodyand dsDNA |
11.47 fM | - | [113] |
| p53gene | QCM** | ssDNA | 1.4pgmL−1 | - | [114] |
*Serum;**Non-Mentioned; AFP: Alpha-Fetoprotein; CA: Cancer Antigen; CEA: Carcinoembryonic Antigen; CRP:C-Reactive Protein; dsDNA: Double-Stranded DNA; EGFR: Epidermal Growth Factor Receptor; PNA: Peptide Nucleic Acid; PSA: Prostate-Specific Antigen; SPR: Surface Plasmon Resonance; ssDNA: Single-Stranded DNA; TNF: Tumor Necrosis Factor; QCM: Quartz Crystal Microbalance.
Table 2 Some current biomarkers and biosensors used in the detection of cancer and their detection limits.
Challenges and Future Prospective
Although there are great advances that have been made in lung cancer biomarker discovery, no data on biomarkers with sufficient sensitivity and specificity has been discovered. This is because of several reasons including: (1) inefficiency know-how applied for biomarker search, (2) distressed reproducibility of laboratory tests, (3) genetic heterogeneity of tumors, (4) an insufficient number of tissue banks for screening, (5) low concentration of analyzed biomarkers and (6) poor research design [99-115].
Many deaths linked with lung cancer have led to several studies directed to the detection of and monitoring the course of the disease. As a result, many lung-specific protein biomarkers have been described that may have the likelihood for diagnosis and prognosis of lung cancer and may play a key role in the pathophysiology of the disease, consequently resulting to better patient care. Hefty challenges still prevail related to reproducibility, marker ratiometric measurements, standardization, and sample handling. Other limitations include; development of new bioinformatics programs that would assist in the analysis of several parameters and samples. Utilizing distinct proteomic platforms may create some disparities in proteomic patterns over different research and clinical centers [116]. The main challenge is found not only in the discovery but in the appropriate evaluation of candidate biomarkers. For instance, a biomarker needs validation with patient samples before entering into clinical practice. Finally, clinical trials would be needed to indicate that these biomarkers are suitable to detect early lung cancers, as early detection decodes enhanced patient survival. The suitability of the biomarkers can be determined by characterizing their sensitivities and specificities and measuring the discriminative cut-off levels. Despite the comprehensive efforts put in these directions, the molecular basis of lung cancer remains still poorly understood to establish the diagnostic, prognostic, and therapeutic importance of biomarkers. Finally, a better understanding of the pathophysiology of lung cancer, various ethical and legal issues also need to be addressed.
Conclusion
In summary, while much improvement has been made in biomarker discovery for early detection of lung cancer, much more still needs to be done because the currently available lung cancer biomarkers are not sufficiently sensitive or specific to be used clinically in diagnosis, stratification, prognosis, or drug responses. As several new potential biomarkers are being scrutinized three facets need to be factored: First, an in-depth analysis of available or potential biomarkers in a large number of clinical samples (including other types of cancer and other disease conditions, like inflammatory diseases) is needed to validate the importance of these biomarkers. In addition, a single biomarker may not be sufficient to envision or monitor the disease; multiplex biomarkers need to be researched to make these helpful for clinical purposes. The biomarker or multiplex biomarkers need to be ratified and get the approval of the Food and Drug Administration (FDA). Finally, more specific and less abundant biomarkers for different subtypes of lung cancers require appropriate attention.
Competing Interests
The authors declare that they have no competing interests
Funding
No funding was given in writing this review paper
Authors' Contributions
DM and PN both substantively revised the manuscript and gave the necessary corrections. LK drafted the manuscript after reading several resources including journals on the area of biomarkers and revised it extensively. All authors read and approved the final manuscript.
Acknowledgements
I would like to acknowledge Damaris Mbui and peter Ndangili for extensively guiding me in writing this manuscript using their expertise in the area of biomarkers. I would like also to thank Damaris Mbui for her encouraging words and moral support.
References
- Bohunicky B, Mousa SA (2010) Biosensors: The new wave in cancer diagnosis. Nanotechnol Sci Appl 4: 1–10.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA et al. (2018a) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6): 394–424.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA et al. (2018b) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6): 394–424.
- Shankar A, Dubey A, Saini D, Singh M, Prasad CP et al. (2019) Environmental and occupational determinants of lung cancer. Transl Lung Cancer Res 8(S1): S31–S49.
- Polanski J, Jankowska-Polanska B, Rosinczuk J, Chabowski M, Szymanska-Chabowska A (2016) Quality of life of patients with lung cancer. Onco Targets Ther 9: 1023–1028.
- Knight SB, Crosbie PA, Balata H, Chudziak J, Hussell T et al. (2017) Progress and prospects of early detection in lung cancer. Open Biology 7(9).
- Zamay TN, Zamay GS, Kolovskaya OS, Zukov RA, Petrova MM et al. (2017) Current and Prospective Protein Biomarkers of Lung Cancer. Cancers 9(11): 155.
- Schnabel DPA, Alllgemeine I (n.d) The new WHO classification of lung cancer (9).
- Dorantes-Heredia R, Ruiz-Morales JM, Cano-García F (2016) Histopathological transformation to small-cell lung carcinoma in non-small cell lung carcinoma tumors. Transl Lung Cancer Res 5(4): 401–412.
- Dai F, Meng S, Mei L, Guan C, Ma Z (2016) Single-port video-assisted thoracic surgery in the treatment of non-small cell lung cancer: A propensity-matched comparative analysis. J Thorac Dis 8(10): 2872–2878.
- Müller KM (1984) Histological classification and histogenesis of lung cancer. Eur J Respir Dis 65(1): 4–19.
- Travis WD (2012) Update on small cell carcinoma and its differentiation from squamous cell carcinoma and other non-small cell carcinomas. Mod Pathol 25 (S1): S18-30.
- Liu Y, Kim J, Qu F, Liu S, Wang H, et al. (2016) CT Features Associated with Epidermal Growth Factor Receptor Mutation Status in Patients with Lung Adenocarcinoma. Radiology 280(1): 271–280.
- LiM, Zhang L, Tang W, Jin YJ, Qi LL, et al. (2019) Identification of epidermal growth factor receptor mutations in pulmonary adenocarcinoma using dual-energy spectral computed tomography. Eur Radiol 29(6):2989–2997.
- Marrugo-Ramírez J, Mir M, Samitier J (2018) Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int. J. Mol. Sci 19(10): 2877.
- Andriani F, Landoni E, Mensah M, Facchinetti F, Miceli R, et al. (2018) Diagnostic role of circulating extracellular matrix-related proteins in non-small cell lung cancer. BMC Cancer 18(1): 899.
- Wojcik E, Kulpa JK (2017) Pro-gastrin-releasing peptide (ProGRP) as a biomarker in small-cell lung cancer diagnosis, monitoring and evaluation of treatment response. Lung Cancer: Targets and Therapy 8: 231–240.
- Gonzalez‐Lopez E, Vrana KE (2020) Dopamine beta-hydroxylase and its genetic variants in human health and disease. J Neurochem, 152(2), 157–181.
- Tian Y, Gong M, Hu Y, Liu H, Zhang W, et al. (2020) Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J Extracell Vesicles 9(1).
- Kosuda A, Shirahata T, Kudo N, Uehara Y, Miyawaki M, et al. (2020) Long-term Survival of a Patient with Small Cell Lung Cancer Secreting ADH and ACTH Simultaneously, Following the Prolonged Use of Amrubicin. Internal Medicine 59(1): 107–112.
- Oh HJ, Park HY, Kim KH, Park CK, Shin HJ, et al. (2016) Progastrin-releasing peptide as a diagnostic and therapeutic biomarker of small cell lung cancer. J Thorac Dis 8(9): 2530–2537.
- Jia H, Zhang L, Wang B (2019) The Value of Combination Analysis of Tumor Biomarkers for Early Differentiating Diagnosis of Lung Cancer and Pulmonary Tuberculosis. Ann Clin Lab Sci 49(5): 645–649.
- Malhotra R, Urs A B, Chakravarti A, Kumar S, Gupta VK, et al. (2016) Correlation of Cyfra 21-1 levels in saliva and serum with CK19 mRNA expression in oral squamous cell carcinoma. Tumor Biology 37(7): 9263–9271.
- Haryati, Diany H (2020) Biomarkers in Lung Cancer. International Journal of Respiratory Medicine 2(2); 11–22.
- Shamji MH, Temblay JN, Cheng W, Byrne SM, Macfarlane E, et al., (2018) Antiapoptotic serine protease inhibitors contribute to survival of allergenic TH2 cells. Journal of Allergy and Clinical Immunology 142(2): 569-581.
- Ye Y, Song YN, He SF, Zhuang JH, Wang GY, et al. (2019) GINS2 promotes cell proliferation and inhibits cell apoptosis in thyroid cancer by regulating CITED2 and LOXL2. Cancer Gene Ther 26(3): 103–113.
- Danielsson F, Peterson MK, Caldeira Araújo H, Lautenschläger F, Gad AKB (2018) Vimentin Diversity in Health and Disease. Cells 7(10): 147.
- Naidoo J, Santos-Zabala ML, Iyriboz T, Woo KM, Sima CS, et al. (2016) Large Cell Neuroendocrine Carcinoma of the Lung: Clinico-Pathologic Features, Treatment, and Outcomes. Clinical Lung Cancer 17(5): e121–e129.
- Nora D, Salluh J, Martin-Loeches I, Póvoa P (2017) Biomarker-guided antibiotic therapy—Strengths and limitations. Ann Transl Med 5(10).
- Bhatnagar S, Katare D, Jain S (2014) Serum-Based Protein Biomarkers for Detection of Lung Cancer. Open Life Sciences 9(4): 341–358.
- Carter JV, Galbraith NJ, Yang D, Burton JF, Walker SP, et al. (2017) Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: A systematic review and meta-analysis. Br J Cancer 116(6): 762–774.
- Wang, Xiaoqian, Kaczor-Urbanowicz KE, Wong DTW (2016) Salivary biomarkers in cancer detection. Medical Oncology 34(1): 7.
- Pu Q, Huang Y, Lu Y, Peng Y, Zhang J, et al. (2016) Tissue-specific and plasma microRNA profiles could be promising biomarkers of histological classification and TNM stage in non-small cell lung cancer. Thoracic Cancer 7(3): 348–354.
- Khamis MM, Adamko DJ, El‐Aneed A (2021) Strategies and Challenges in Method Development and Validation for the Absolute Quantification of Endogenous Biomarker Metabolites Using Liquid Chromatography-Tandem Mass Spectrometry. Mass Spectrometry Reviews 40(1): 31–52.
- Olkowicz M, Debski J, Jablonska P, Dadlez M, Smolenski RT (2017) Application of a new procedure for liquid chromatography/mass spectrometry profiling of plasma amino acid-related metabolites and untargeted shotgun proteomics to identify mechanisms and biomarkers of calcific aortic stenosis. J Chromatography A 1517: 66–78.
- Sobsey CA, Ibrahim S, Richard VR, Gaspar V, Mitsa G, et al. (2020) Targeted and Untargeted Proteomics Approaches in Biomarker Development. Proteomics 20(9).
- Paczesny S (2018) Biomarkers for posttransplantation outcomes. Blood 131(20): 2193–2204.
- Sharma D, Farahbakhsh N, Shastri S, Sharma P (2018) Biomarkers for diagnosis of neonatal sepsis: A literature review. The Journal of Maternal-Fetal & Neonatal Medicine 31(12): 1646–1659.
- Whitwell JL, Höglinger GU, Antonini A, Bordelon Y, Boxer AL, et al. (2017) Radiological biomarkers for diagnosis in PSP: Where are we and where do we need to be? Movement Disorders 32(7): 955–971.
- Turner CT, Gupta RK, Tsaliki E, Roe JK, Mondal P, et al. (2020) Blood transcriptional biomarkers for active pulmonary tuberculosis in a high-burden setting: A prospective, observational, diagnostic accuracy study. The Lancet Respiratory Medicine 8(4): 407– 419.
- Sadighbayan D, Sadighbayan K, Tohid-kia MR, Khosroushahi AY, Hasanzadeh M (2019) Development of electrochemical biosensors for tumor marker determination towards cancer diagnosis: Recent progress. TrAC Trends in Analytical Chemistry, 118: 73–88.
- Huang R, He N, Li Z (2018) Recent progresses in DNA nanostructure-based biosensors for detection of tumor markers. Biosensors and Bioelectronics 109: 27–34.
- Gashimova EM, Temerdashev AZ, Porkhanov VA, Polyakov IS, Perunov DV, et al. (2019) Evaluation of the Possibility of Volatile Organic Compounds Determination in Exhaled Air by Gas Chromatography for the Noninvasive Diagnostics of Lung Cancer. J Analytical Chemistry 74(5): 472– 479.
- Tarro G, Paolini M, Rossi A (2019) Molecular Biology of Lung Cancer and Future Perspectives for Screening. Mass Spectrometry - Future Perceptions and Applications.
- Llop E, Guerrero PE, Duran A, Barrabés S, Massaguer A, et al., (2018). Glycoprotein biomarkers for the detection of pancreatic ductal adenocarcinoma. World J Gastroenterol 24(24): 2537.
- Jelski W, Mroczko B (2020) Biochemical Markers of Colorectal Cancer – Present and Future. Cancer Management and Research 12: 4789.
- Chen C, Chen Q, Zhao Q, Liu M, Guo J (2017) Value of Combined Detection of Serum CEA, CA72-4, CA19-9, CA15-3 and CA12-5 in the Diagnosis of Gastric Cancer. Ann Clin Lab Sci 47(3): 260–263.
- Hackner K, Errhalt P, Handzhiev S (2019) Ratio of carcinoembryonic antigen in pleural fluid and serum for the diagnosis of malignant pleural effusion. Therapeutic Advances in Medical Oncology (11).
- Zhang H, Li C, Hu F, Zhang X, Shen Y, et al. (2020) Auxiliary diagnostic value of tumor biomarkers in pleural fluid for lung cancer-associated malignant pleural effusion. Respir Res 21(1): 284.
- Korkmaz ET, Koksal D, Aksu F, Dikmen ZG, Icen D, et al. (2018) Triple test with tumor markers CYFRA 21.1, HE4, and ProGRP might contribute to diagnosis and subtyping of lung cancer. Clinical Biochemistry 58: 15–19.
- Tang KD, Kenny L, Perry C, Frazer I, Punyadeera C (2017) The overexpression of salivary cytokeratins as potential diagnostic biomarkers in head and neck squamous cell carcinomas. Oncotarget 8(42): 72272–72280.
- Yang G, Xiao Z, Tang C, Deng Y, Huang H, et al. (2019) Recent advances in biosensor for detection of lung cancer biomarkers. Biosensors and Bioelectronics 141: 111416.
- Fu L, Wang R, Yin L, Shang X, Zhang R, et al. (2019) CYFRA21-1 tests in the diagnosis of non-small cell lung cancer: A meta-analysis. The International J Biological Markers 34(3): 251–261.
- Chen M, Wang Y, Su H, Mao L, Jiang X, et al. (2018) Three-dimensional electrochemical DNA biosensor based on 3D graphene-Ag nanoparticles for sensitive detection of CYFRA21-1 in non-small cell lung cancer. Sensors and Actuators B: Chemical 255: 2910–2918.
- Fu YJ, Li KZ, Bai JH, Liang ZQ (2019) C-reactive protein/albumin ratio is a prognostic indicator in Asians with pancreatic cancers. Medicine 98(48).
- Wang L, Hu R, Liu H, Li W, Zhou L, et al. (2020) Potentials of neuron- specific enolase as a biomarker for gastric cancer. Tropical Journal of Pharmaceutical Research 19(3): 505–511.
- Rzechonek A, Kaminska A, Mamczur P, Drapiewski A, Budzynski W (2017) Limited Clinical Significance of Dimeric Form of Pyruvate Kinase as a Diagnostic and Prognostic Biomarker in Non-small Cell Lung Cancer. In M. Pokorski (Ed.), Pathobiology of Pulmonary Disorders (pp. 51–57).
- Bandyopadhayaya S, Mandal CC (2020) Blood-Based Biomarkers for the Diagnosis and Prognosis of Cancer. In Precision Medicine in Oncology (pp. 61–82).
- Zhang XY, Zhang G, Jiang Y, Liu D, Li MZ, et al. (2015) The prognostic value of serum C-reactive protein-bound serum amyloid A in early-stage lung cancer. Chin J Cancer 34(3): 1–15.
- Sproston NR, Ashworth JJ (2018) Role of C-Reactive Protein at Sites of Inflammation and Infection. Frontiers in Immunology (9).
- McFadyen JD, Kiefer J, Braig D, Loseff-Silver J, Potempa LA, et al. (2018) Dissociation of C-Reactive Protein Localizes and Amplifies Inflammation: Evidence for a Direct Biological Role of C-Reactive Protein and Its Conformational Changes. Frontiers in Immunology (9)
- Nakayama T, Saito K, Kumagai J, Nakajima Y, Kijima T, et al. (2018) Higher Serum C-reactive Protein Level Represents the Immunosuppressive Tumor Microenvironment in Patients With Clear Cell Renal Cell Carcinoma. Clinical Genitourinary Cancer 16(6): e1151–e1158.
- Meyer MM, Brandenburg L, Hudel H, Agné A, Padberg W, et al. (2020) Who Is Afraid of CRP? Elevated Preoperative CRP Levels Might Attenuate the Increase in Inflammatory Parameters in Response to Lung Cancer Surgery. J Clin Med 9(10): 3340.
- Li Z, Li Y, Fu J, Li N, Shen L (2019) Clinical utility of microRNA-451 as diagnostic biomarker for human cancers. Bioscience Reports 39(1).
- Schlick K, Magnes T, Huemer F, Ratzinger L, Weiss L, et al. (2019) C-Reactive Protein and Neutrophil/Lymphocytes Ratio: Prognostic Indicator for Doubling Overall Survival Prediction in Pancreatic Cancer Patients. J Clin Med 8(11): 1791.
- Zhou L, Cai X, Liu Q, Jian ZY, Li H et al. (2015) Prognostic Role of C- Reactive Protein In Urological Cancers: A Meta-Analysis. Scientific Reports 5(1): 12733.
- Gan W, Yi Y, Fu Y, Huang J, Lu Z, et al. (2018) Fibrinogen and C-reactive protein score is a prognostic index for patients with hepatocellular carcinoma undergoing curative resection: A prognostic nomogram study. J Cancer 9(1): 148–156.
- Yamamoto M, Saito H, Uejima C, Tanio A, Takaya S, et al. (2018) Prognostic value of the combination of pre- and postoperative C-reactive protein in colorectal cancer patients. Surgery Today 48(11): 986–993.
- Shibutani M, Maeda K, Nagahara H, Iseki Y, Ikeya T, Hirakawa K (2016) Prognostic Significance of the Preoperative Ratio of C-Reactive Protein to Albumin in Patients with Colorectal Cancer. Anticancer Research, 36(3), 995–1001.
- Yang JR, Xu JY, Chen GC, Yu N, Yang J,et al., (2019) Post-diagnostic C-reactive protein and albumin predict survival in Chinese patients with non-small cell lung cancer: A prospective cohort study. Scientific Reports 9(1): 8143.
- Xiao X, Wang S, Long G (2019) C-reactive protein is a significant predictor of improved survival in patients with advanced non-small cell lung cancer. Medicine 98(26).
- Wang Xiaolin, Liu S, Zhao X, Fang E, Zhao, X (2019) The value of C-reactive protein as an independent prognostic indicator for disease-specific survival in patients with soft tissue sarcoma: A meta-analysis. PLOS ONE, 14(7).
- Chi PD, Liu W, Chen H, Zhang JP, Lin Y, et al., (2014) High- density lipoprotein cholesterol is a favorable prognostic factor and negatively correlated with C-reactive protein level in non-small cell lung carcinoma. PloS One, 9(3).
- Wu J, Shen Z (2020) Exosomal miRNAs as biomarkers for diagnostic and prognostic in lung cancer. Cancer Medicine 9(19): 6909–6922.
- Yáñez-Mó M, Siljander PRM, Andreu Z, Zavec AB, Borràs FE, et al., (2015) Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 4: 27066.
- Merchant ML, Rood IM, Deegens JKJ, Klein JB (2017) Isolation and characterization of urinary extracellular vesicles: Implications for biomarker discovery. Nature Reviews. Nephrology 13(12): 731–749.
- Zhang Q, Liu Y, Nie Y, Liu Y, Ma Q (2019) Wavelength-Dependent Surface Plasmon Coupling Electrochemiluminescence Biosensor Based on Sulfur-Doped Carbon Nitride Quantum Dots for K-RAS Gene Detection. Anal Chem. 91(21): 13780–13786.
- Lin Y, Dong H, Deng W, Lin W, Li K, et al., (2019) Evaluation of Salivary Exosomal Chimeric GOLM1-NAA35 RNA as a Potential Biomarker in Esophageal Carcinoma. Clinical Cancer Research 25(10): 3035–3045.
- Severino V, Dumonceau JM, Delhaye M, Moll S, Annessi-Ramseyer I, et al., (2017) Extracellular Vesicles in Bile as Markers of Malignant Biliary Stenoses. Gastroenterology 153(2): 495-504.
- Qin W, Tsukasaki Y, Dasgupta S, Mukhopadhyay N, Ikebe M et al., (2016) Exosomes in Human Breast Milk Promote EMT. Clin Cancer Res 22(17): 4517–4524.
- Xiao GY, Cheng CC, Chiang YS, Cheng WTK, Liu IH, et al., (2016) Exosomal miR-10a derived from amniotic fluid stem cells preserves ovarian follicles after chemotherapy. Scientific Reports.
- Li G, Li S, Wang Z, Xue Y, Dong, C, et al., (2018). Label-free electrochemical aptasensor for detection of alpha-fetoprotein based on AFP- aptamer and thionin/reduced graphene oxide/gold nanoparticles. Anal Biochem, 547: 37–44.
- TO, Yagi YHK, AM, HM, SG, et al., (2017) Next- generation sequencing-based small RNA profiling of cerebrospinal fluid exosomes. Neuroscience Letters; Neurosci Lett.
- Tian Z, Liang C, Zhang Z, Wen H, Feng H, et al., (2020) Prognostic value of neuron-specific enolase for small cell lung cancer: A systematic review and meta-analysis. World Journal of Surgical Oncology 18(1): 1–8.
- Ando W, Kikuchi K, Uematsu T, Yokomori H, Takaki T, et al., (2019). Novel breast cancer screening: Combined expression of miR-21 and MMP-1 in urinary exosomes detects 95% of breast cancer without metastasis. Scientific Reports 9(1): 13595.
- Zhou S, Hu T, Zhang F, Tang, D, Li D, et al., (2020) Integrated Microfluidic Device for Accurate Extracellular Vesicle Quantification and Protein Markers Analysis Directly from Human Whole Blood. Anal Chem 92(1): 1574–1581.
- Martins TS, Catita J, Rosa IM, Silva OABdaCe, Henriques AG (2018) Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLOS ONE 13(6).
- Rodríguez M, Silva J, López-Alfonso A, López-Muñiz MB, Peña C, et al., (2014) Different exosome cargo from plasma/bronchoalveolar lavage in non-small-cell lung cancer. Genes, Chromosomes & Cancer 53(9): 713–724.
- Clayton A, Boilard E, Buzas EI, Cheng L, Falcón-Perez JM (2019) Considerations towards a roadmap for collection, handling and storage of blood extracellular vesicles. J Extracell Vesicles 8(1): 1647027.
- Wen G, Zhou T, Gu W (2020) The potential of using blood circular RNA as liquid biopsy biomarker for human diseases. Protein & Cell.
- Hofmann L, Ludwig S, Vahl JM, Brunner C, Hoffmann TK, et al., (2020) The Emerging Role of Exosomes in Diagnosis, Prognosis, and Therapy in Head and Neck Cancer. International Journal of Molecular Sciences 21(11): 4072.
- Lv J, Zhao HP, Dai K, Cheng, Y, Zhang J, et al., (2020) Circulating exosomal miRNAs as potential biomarkers for Barrett’s esophagus and esophageal adenocarcinoma. World Journal of Gastroenterology 26(22): 2889–2901.
- Syn N, Wang L, Sethi G, Thiery JP, Goh BC (2016) Exosome-Mediated Metastasis: From Epithelial-Mesenchymal Transition to Escape from Immunosurveillance. Trends in Pharmacological Sciences 37(7): 606–617.
- Hsu YL, Hung JY, Chang WA, Lin YS, Pan YC, et al., (2017) Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO- 1. Oncogene 36(34): 4929–4942.
- Lobb RJ, van Amerongen R, Wiegmans A, Ham S, Larsen JE, et al., (2017) Exosomes derived from mesenchymal non-small cell lung cancer cells promote chemoresistance. International Journal of Cancer 141(3): 614–620.
- Turchinovich A, Tonevitsky AG, Cho WC, Burwinkel B (2015) Check and mate to exosomal extracellular miRNA: New lesson from a new approach. Frontiers in Molecular Biosciences (2).
- Wu D, Deng S, Liu T, Han R, Zhang T, et al., (2018) TGF-β-mediated exosomal lnc- MMP2-2 regulates migration and invasion of lung cancer cells to the vasculature by promoting MMP2 expression. Cancer Medicine (7).
- Dejima H, Iinuma H, Kanaoka R, Matsutani N, Kawamura M (2017) Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncology Letters 13(3): 1256–1263.
- Regiart M, Fernández-Baldo MA, Villarroel-Rocha J, Messina GA, Bertolino FA, et al., (2017) Microfluidic immunosensor based on mesoporous silica platform and CMK-3/poly-acrylamide-co-methacrylate of dihydrolipoic acid modified gold electrode for cancer biomarker detection. Analytica Chimica Acta 963: 83–92.
- Li S, Wan Y, Su Y, Fan C, Bhethanabotla VR (2017) Gold nanoparticle-based low limit of detection Love wave biosensor for carcinoembryonic antigens. Biosensors and Bioelectronics 95: 48–54.
- Wang J,Hui N (2019) Zwitterionic poly(carboxybetaine) functionalized conductingpolymerpolyanilinenanowiresfortheelectrochemicaldetectionof carcinoembryonicantigeninundilutedbloodserum.Bioelectrochemistry125:90–96.
- Ermini ML, Chadtová Song X, Špringer T, Homola J (2019) Peptide Functionalization of Gold Nanoparticles for the Detection of Carcinoembryonic Antigen in Blood Plasma via SPR-Based Biosensor. Frontiers in Chemistry (7).
- Jeng MJ, Sharma M, Li YC, Lu YC, Yu CY, et al., (2020) Surface Acoustic Wave Sensor for C-Reactive Protein Detection. Sensors 20(22): 6640.
- Singh MK, Pal S, Verma A, Prajapati YK, Saini JP (2020) Highly sensitive antimonene-coated black phosphorous-based surface plasmon-resonance biosensor for DNA hybridization: Design and numerical analysis. Journal of Nanophotonics 14(4): 046015.
- Akbari Nakhjavani S, Khalilzadeh B, Samadi Pakchin P, Saber R, Ghahremani MH, et al., (2018) A highly sensitive and reliable detection of CA15-3 in patient plasma with electrochemical biosensor labeled with magnetic beads. Biosensors and Bioelectronics 122: 8–15.
- Wang H, Wang X, Wang J, Fu W, Yao C (2016) A SPR biosensor based on signal amplification using antibody-QD conjugates for quantitative determination of multiple tumor markers. Scientific Reports 6(1): 33140.
- Chinnadayyala SR, Park J, Kim YH, Choi SH, Lee SM, et al., (2019) Electrochemical Detection of C-Reactive Protein in Human Serum Based on Self-Assembled Monolayer-Modified Interdigitated Wave- Shaped Electrode. Sensors 19(24): 5560.
- Ghosh S, Datta D, Chaudhry S, Dutta M, Stroscio MA (2018) Rapid Detection of Tumor Necrosis Factor-Alpha Using Quantum Dot-Based Optical Aptasensor. IEEE Transactions on NanoBioscience, 17(4), 417–423.
- Ranganathan V, Srinivasan S, Singh A, DeRosa MC (2020) An aptamer-based colorimetric lateral flow assay for the detection of human epidermal growth factor receptor 2 (HER2). Analytical Biochemistry 588: 113471.
- Johari-Ahar M, Karami P, Ghanei M, Afkhami A, Bagheri H (2018) Development of a molecularly imprinted polymer tailored on disposable screen-printed electrodes for dual detection of EGFR and VEGF using nano-liposomal amplification strategy. Biosensors and Bioelectronics 107: 26–33.
- Jonous ZA, Shayeh JS, Yazdian F, Yadegari A, Hashemi M, et al., (2019) An electrochemical biosensor for prostate cancer biomarker detection using graphene oxide– gold nanostructures. Engineering in Life Sciences 19(3): 206–216.
- Ertürk G, Özen H, Tümer MA, Mattiasson B, Denizli A (2016) Microcontact imprinting based surface plasmon resonance (SPR) biosensor for real-time and ultrasensitive detection of prostate specific antigen (PSA) from clinical samples. Sensors and Actuators B: Chemical 224: 823–832.
- Song S, Lee JU, Kang J, Park KH, Sim SJ (2020) Real-time monitoring of distinct binding kinetics of hot-spot mutant p53 protein in human cancer cells using an individual nanorod-based plasmonic biosensor. Sensors and Actuators B: Chemical 322: 128584.
- Soares JC, Soares AC, Pereira PAR, Rodrigues VdaC, Shimizu FM, et al., (2016). Adsorption according to the Langmuir–Freundlich model is the detection mechanism of the antigen p53 for early diagnosis of cancer. Physical Chemistry Chemical Physics 18(12): 8412–8418.
- Ren AH, Fiala CA, Diamandis EP, Kulasingam V (2020) Pitfalls in Cancer Biomarker Discovery and Validation with Emphasis on Circulating Tumor DNA. Cancer Epidemiology and Prevention Biomarkers 29(12): 2568–2574.
- Hristova VA, Chan DW (2019) Cancer biomarker discovery and translation: Proteomics and beyond. Expert Review of Proteomics 16(2): 93–103.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences